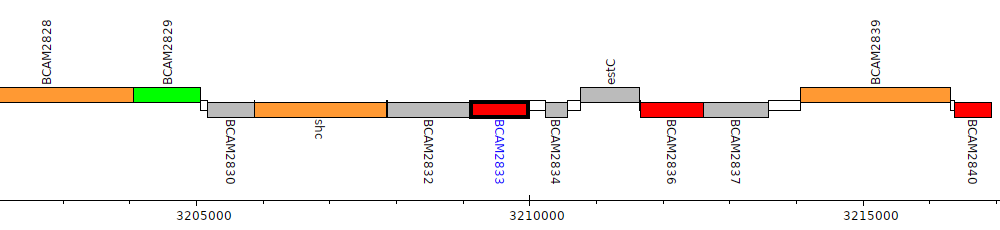
Burkholderia cenocepacia J2315, BCAM2833
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0016117 | carotenoid biosynthetic process |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR03465
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups |
Inferred from Sequence Model
Term mapped from: InterPro:PS01045
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity |
Inferred from Sequence Model
Term mapped from: InterPro:cd00683
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0016767 | geranylgeranyl-diphosphate geranylgeranyltransferase activity |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR03465
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0006696 | ergosterol biosynthetic process |
Inferred from Sequence Model
Term mapped from: InterPro:cd00683
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0051996 | squalene synthase activity |
Inferred from Sequence Model
Term mapped from: InterPro:cd00683
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | bcj00906 | Carotenoid biosynthesis | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | bcj01100 | Metabolic pathways | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | bcj01110 | Biosynthesis of secondary metabolites | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| SUPERFAMILY | SSF48576 | IPR008949 | Isoprenoid synthase domain superfamily | 20 | 281 | 1.87E-62 | |
| TIGRFAM | TIGR03465 | HpnD: squalene synthase HpnD | IPR017828 | Squalene synthase HpnD-like | 17 | 281 | 7.3E-101 |
| Pfam | PF00494 | Squalene/phytoene synthase | 20 | 266 | 8.4E-59 | ||
| ProSitePatterns | PS01045 | Squalene and phytoene synthases signature 2. | IPR019845 | Squalene/phytoene synthase, conserved site | 158 | 183 | - |
| Gene3D | G3DSA:1.10.600.10 | IPR008949 | Isoprenoid synthase domain superfamily | 7 | 260 | 1.2E-63 | |
| CDD | cd00683 | Trans_IPPS_HH | IPR033904 | Trans-Isoprenyl Diphosphate Synthases, head-to-head | 20 | 272 | 1.15071E-72 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.