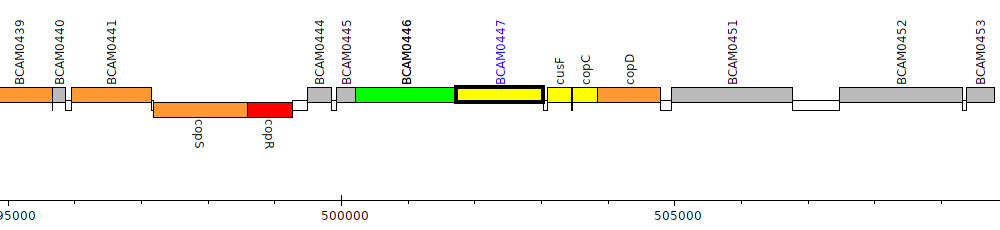
Burkholderia cenocepacia J2315, BCAM0447
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0016491 | oxidoreductase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PF07731
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0005507 | copper ion binding |
Inferred from Sequence Model
Term mapped from: InterPro:PF07731
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0055114 | oxidation-reduction process |
Inferred from Sequence Model
Term mapped from: InterPro:PF07731
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| ProSiteProfiles | PS51318 | Twin arginine translocation (Tat) signal profile. | IPR006311 | Twin-arginine translocation pathway, signal sequence | 1 | 26 | 8.768 |
| Pfam | PF07732 | Multicopper oxidase | IPR011707 | Multicopper oxidase, type 3 | 84 | 193 | 1.0E-38 |
| Pfam | PF07731 | Multicopper oxidase | IPR011706 | Multicopper oxidase, type 2 | 217 | 332 | 6.3E-24 |
| Gene3D | G3DSA:2.60.40.420 | IPR008972 | Cupredoxin | 64 | 333 | 3.2E-77 | |
| ProSitePatterns | PS00080 | Multicopper oxidases signature 2. | IPR002355 | Multicopper oxidase, copper-binding site | 316 | 327 | - |
| CDD | cd04202 | CuRO_D2_2dMcoN_like | 200 | 337 | 2.9444E-65 | ||
| SUPERFAMILY | SSF49503 | IPR008972 | Cupredoxin | 43 | 193 | 7.18E-40 | |
| SUPERFAMILY | SSF49503 | IPR008972 | Cupredoxin | 172 | 329 | 2.7E-35 | |
| CDD | cd13860 | CuRO_1_2dMco_1 | 72 | 190 | 1.33715E-74 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.