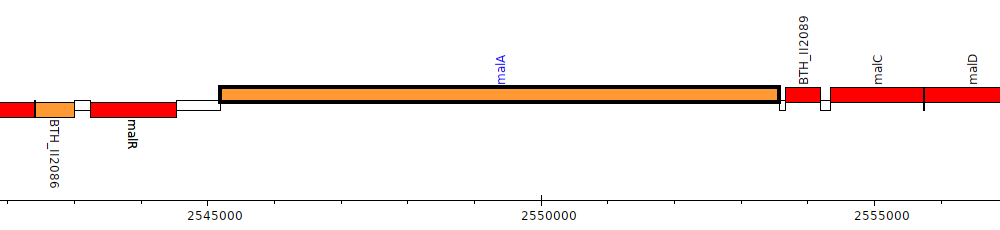
Burkholderia thailandensis E264 ATCC 700388, BTH_II2088 (malA)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0016788 | hydrolase activity, acting on ester bonds |
Inferred from Sequence Model
Term mapped from: InterPro:SM00824
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0031177 | phosphopantetheine binding |
Inferred from Sequence Model
Term mapped from: InterPro:SM00823
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0009058 | biosynthetic process |
Inferred from Sequence Model
Term mapped from: InterPro:SM00824
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0003824 | catalytic activity |
Inferred from Sequence Model
Term mapped from: InterPro:G3DSA:3.40.47.10
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0016740 | transferase activity |
Inferred from Sequence Model
Term mapped from: InterPro:G3DSA:3.40.366.10
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Pfam | PF00975 | Thioesterase domain | IPR001031 | Thioesterase | 1947 | 2167 | 5.1E-13 |
| Gene3D | G3DSA:3.40.47.10 | IPR016039 | Thiolase-like | 909 | 1359 | 2.8E-141 | |
| SMART | SM00823 | Phosphopantetheine attachment site | IPR020806 | Polyketide synthase, phosphopantetheine-binding domain | 817 | 886 | 3.5E-4 |
| CDD | cd00833 | PKS | 914 | 1354 | 1.21002E-156 | ||
| SMART | SM00825 | Beta-ketoacyl synthase | IPR020841 | Polyketide synthase, beta-ketoacyl synthase domain | 914 | 1359 | 3.4E-139 |
| SUPERFAMILY | SSF51735 | IPR036291 | NAD(P)-binding domain superfamily | 2191 | 2373 | 5.9E-10 | |
| ProSitePatterns | PS00012 | Phosphopantetheine attachment site. | IPR006162 | Phosphopantetheine attachment site | 2741 | 2756 | - |
| Pfam | PF02801 | Beta-ketoacyl synthase, C-terminal domain | IPR014031 | Beta-ketoacyl synthase, C-terminal | 1173 | 1298 | 1.4E-34 |
| Gene3D | G3DSA:3.40.50.1820 | IPR029058 | Alpha/Beta hydrolase fold | 1911 | 2180 | 2.6E-27 | |
| Gene3D | G3DSA:1.10.1200.10 | IPR036736 | ACP-like superfamily | 800 | 889 | 1.6E-21 | |
| Gene3D | G3DSA:3.30.70.3290 | 1375 | 1838 | 5.5E-107 | |||
| Pfam | PF00109 | Beta-ketoacyl synthase, N-terminal domain | IPR014030 | Beta-ketoacyl synthase, N-terminal | 913 | 1164 | 4.7E-59 |
| SUPERFAMILY | SSF53474 | IPR029058 | Alpha/Beta hydrolase fold | 13 | 250 | 4.88E-42 | |
| Pfam | PF16197 | Ketoacyl-synthetase C-terminal extension | IPR032821 | Ketoacyl-synthetase, C-terminal extension | 1331 | 1421 | 1.5E-13 |
| ProSitePatterns | PS00012 | Phosphopantetheine attachment site. | IPR006162 | Phosphopantetheine attachment site | 841 | 856 | - |
| ProSitePatterns | PS00455 | Putative AMP-binding domain signature. | IPR020845 | AMP-binding, conserved site | 434 | 445 | - |
| SMART | SM00822 | This enzymatic domain is part of bacterial polyketide synthases and catalyses the first step in the reductive modification of the beta-carbonyl centres in the growing polyketide chain. It uses NADPH to reduce the keto group to a hydroxy group. | 2387 | 2553 | 1.3E-18 | ||
| Gene3D | G3DSA:3.30.300.30 | 692 | 799 | 5.9E-20 | |||
| Pfam | PF00550 | Phosphopantetheine attachment site | IPR009081 | Phosphopantetheine binding ACP domain | 2738 | 2774 | 2.6E-4 |
| MobiDBLite | mobidb-lite | consensus disorder prediction | 789 | 812 | - | ||
| Pfam | PF08659 | KR domain | IPR013968 | Polyketide synthase, ketoreductase domain | 2387 | 2552 | 2.2E-31 |
| SUPERFAMILY | SSF55048 | IPR016036 | Malonyl-CoA ACP transacylase, ACP-binding | 1604 | 1667 | 1.7E-8 | |
| SUPERFAMILY | SSF47336 | IPR036736 | ACP-like superfamily | 815 | 882 | 1.57E-15 | |
| Gene3D | G3DSA:3.40.366.10 | IPR001227 | Acyl transferase domain superfamily | 1467 | 1788 | 5.5E-107 | |
| SUPERFAMILY | SSF56801 | 287 | 824 | 1.44E-102 | |||
| ProSitePatterns | PS00606 | Beta-ketoacyl synthases active site. | IPR018201 | Beta-ketoacyl synthase, active site | 1079 | 1095 | - |
| SUPERFAMILY | SSF51735 | IPR036291 | NAD(P)-binding domain superfamily | 2386 | 2569 | 2.05E-26 | |
| Pfam | PF00975 | Thioesterase domain | IPR001031 | Thioesterase | 17 | 240 | 9.6E-38 |
| CDD | cd05930 | A_NRPS | 301 | 793 | 2.67058E-109 | ||
| MobiDBLite | mobidb-lite | consensus disorder prediction | 2660 | 2677 | - | ||
| SMART | SM00824 | Thioesterase | IPR020802 | Polyketide synthase, thioesterase domain | 19 | 251 | 1.3E-5 |
| SUPERFAMILY | SSF47336 | IPR036736 | ACP-like superfamily | 2720 | 2782 | 1.7E-5 | |
| Gene3D | G3DSA:3.40.50.12780 | IPR042099 | AMP-dependent synthetase-like superfamily | 280 | 691 | 2.2E-85 | |
| SUPERFAMILY | SSF53901 | IPR016039 | Thiolase-like | 910 | 1354 | 1.87E-72 | |
| Gene3D | G3DSA:3.40.50.720 | 2181 | 2630 | 9.6E-61 | |||
| ProSiteProfiles | PS50075 | Carrier protein (CP) domain profile. | IPR009081 | Phosphopantetheine binding ACP domain | 811 | 886 | 15.652 |
| Pfam | PF13193 | AMP-binding enzyme C-terminal domain | IPR025110 | AMP-binding enzyme, C-terminal domain | 707 | 777 | 2.2E-6 |
| Pfam | PF00698 | Acyl transferase domain | IPR014043 | Acyl transferase | 1468 | 1758 | 3.6E-39 |
| SUPERFAMILY | SSF53474 | IPR029058 | Alpha/Beta hydrolase fold | 1920 | 2178 | 1.53E-22 | |
| Gene3D | G3DSA:3.40.50.1820 | IPR029058 | Alpha/Beta hydrolase fold | 2 | 248 | 3.3E-55 | |
| SMART | SM00827 | Acyl transferase domain in polyketide synthase (PKS) enzymes. | IPR020801 | Polyketide synthase, acyl transferase domain | 1469 | 1782 | 5.5E-78 |
| Pfam | PF00501 | AMP-binding enzyme | IPR000873 | AMP-dependent synthetase/ligase | 294 | 698 | 3.2E-67 |
| SUPERFAMILY | SSF52151 | IPR016035 | Acyl transferase/acyl hydrolase/lysophospholipase | 1466 | 1758 | 4.45E-57 | |
| Pfam | PF00550 | Phosphopantetheine attachment site | IPR009081 | Phosphopantetheine binding ACP domain | 819 | 881 | 7.2E-13 |
| MobiDBLite | mobidb-lite | consensus disorder prediction | 2630 | 2716 | - |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.