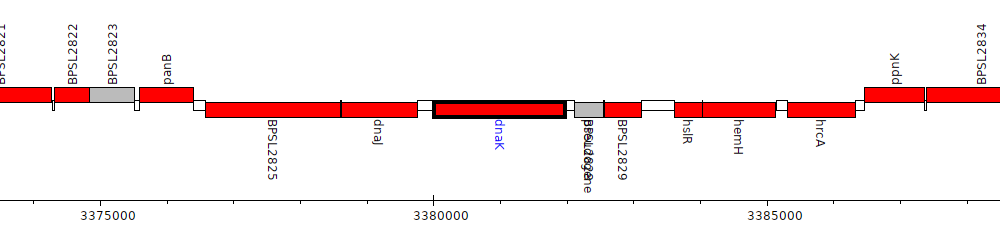
Burkholderia pseudomallei K96243, BPSL2827 (dnaK)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0006457 | protein folding |
Inferred from Sequence Model
Term mapped from: InterPro:MF_00332
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0005524 | ATP binding |
Inferred from Sequence Model
Term mapped from: InterPro:MF_00332
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0051082 | unfolded protein binding |
Inferred from Sequence Model
Term mapped from: InterPro:MF_00332
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | bps03018 | RNA degradation | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Gene3D | G3DSA:3.90.640.10 | 230 | 317 | 3.8E-165 | |||
| Gene3D | G3DSA:3.30.420.40 | 180 | 367 | 3.8E-165 | |||
| Gene3D | G3DSA:1.20.1270.10 | IPR029048 | Heat shock protein 70kD, C-terminal domain superfamily | 525 | 610 | 1.4E-23 | |
| PRINTS | PR00301 | 70kDa heat shock protein signature | IPR013126 | Heat shock protein 70 family | 138 | 158 | 2.2E-80 |
| ProSitePatterns | PS00329 | Heat shock hsp70 proteins family signature 2. | IPR018181 | Heat shock protein 70, conserved site | 193 | 206 | - |
| PRINTS | PR00301 | 70kDa heat shock protein signature | IPR013126 | Heat shock protein 70 family | 470 | 486 | 2.2E-80 |
| PRINTS | PR00301 | 70kDa heat shock protein signature | IPR013126 | Heat shock protein 70 family | 366 | 386 | 2.2E-80 |
| SUPERFAMILY | SSF100920 | IPR029047 | Heat shock protein 70kD, peptide-binding domain superfamily | 385 | 540 | 1.7E-60 | |
| SUPERFAMILY | SSF53067 | 188 | 383 | 2.22E-69 | |||
| Pfam | PF00012 | Hsp70 protein | IPR013126 | Heat shock protein 70 family | 4 | 606 | 2.2E-263 |
| Gene3D | G3DSA:2.60.34.10 | IPR029047 | Heat shock protein 70kD, peptide-binding domain superfamily | 384 | 524 | 9.5E-67 | |
| Coils | Coil | 260 | 280 | - | |||
| Gene3D | G3DSA:3.30.420.40 | 5 | 380 | 3.8E-165 | |||
| ProSitePatterns | PS00297 | Heat shock hsp70 proteins family signature 1. | IPR018181 | Heat shock protein 70, conserved site | 7 | 14 | - |
| PRINTS | PR00301 | 70kDa heat shock protein signature | IPR013126 | Heat shock protein 70 family | 389 | 408 | 2.2E-80 |
| TIGRFAM | TIGR02350 | prok_dnaK: chaperone protein DnaK | IPR012725 | Chaperone DnaK | 3 | 606 | 5.6E-288 |
| SUPERFAMILY | SSF53067 | 4 | 183 | 3.08E-63 | |||
| ProSitePatterns | PS01036 | Heat shock hsp70 proteins family signature 3. | IPR018181 | Heat shock protein 70, conserved site | 338 | 352 | - |
| SUPERFAMILY | SSF100934 | IPR029048 | Heat shock protein 70kD, C-terminal domain superfamily | 508 | 607 | 6.02E-20 | |
| MobiDBLite | mobidb-lite | consensus disorder prediction | 611 | 650 | - | ||
| PRINTS | PR00301 | 70kDa heat shock protein signature | IPR013126 | Heat shock protein 70 family | 199 | 209 | 2.2E-80 |
| PRINTS | PR00301 | 70kDa heat shock protein signature | IPR013126 | Heat shock protein 70 family | 54 | 62 | 2.2E-80 |
| Hamap | MF_00332 | Chaperone protein DnaK [dnaK]. | IPR012725 | Chaperone DnaK | 1 | 647 | 36.863 |
| Coils | Coil | 563 | 590 | - | |||
| PRINTS | PR00301 | 70kDa heat shock protein signature | IPR013126 | Heat shock protein 70 family | 335 | 351 | 2.2E-80 |
| PRINTS | PR00301 | 70kDa heat shock protein signature | IPR013126 | Heat shock protein 70 family | 3 | 16 | 2.2E-80 |
| PRINTS | PR00301 | 70kDa heat shock protein signature | IPR013126 | Heat shock protein 70 family | 31 | 43 | 2.2E-80 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.