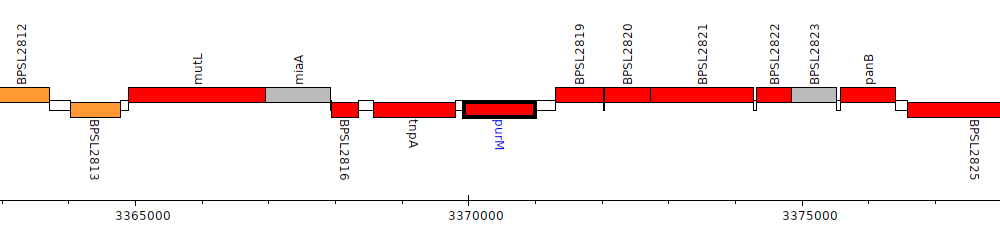
Burkholderia pseudomallei K96243, BPSL2818 (purM)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0006189 | 'de novo' IMP biosynthetic process |
Inferred from Sequence Model
Term mapped from: InterPro:cd02196
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0004641 | phosphoribosylformylglycinamidine cyclo-ligase activity |
Inferred from Sequence Model
Term mapped from: InterPro:cd02196
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| MetaCyc | 5-aminoimidazole ribonucleotide biosynthesis I | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
||
| KEGG | bps01110 | Biosynthesis of secondary metabolites | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | bps01100 | Metabolic pathways | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | bps00230 | Purine metabolism | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| MetaCyc | superpathway of 5-aminoimidazole ribonucleotide biosynthesis | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
||
| KEGG (InterPro) | 00230 | Purine metabolism | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
|
| KEGG | bps01130 | Biosynthesis of antibiotics | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| MetaCyc | 5-aminoimidazole ribonucleotide biosynthesis II | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Gene3D | G3DSA:3.90.650.10 | IPR036676 | PurM-like, C-terminal domain superfamily | 178 | 351 | 1.3E-70 | |
| SUPERFAMILY | SSF55326 | IPR036921 | PurM-like, N-terminal domain superfamily | 13 | 176 | 1.2E-53 | |
| Pfam | PF00586 | AIR synthase related protein, N-terminal domain | IPR016188 | PurM-like, N-terminal domain | 70 | 170 | 2.3E-15 |
| Gene3D | G3DSA:3.30.1330.10 | IPR036921 | PurM-like, N-terminal domain superfamily | 12 | 176 | 8.3E-69 | |
| CDD | cd02196 | PurM | IPR004733 | Phosphoribosylformylglycinamidine cyclo-ligase | 53 | 338 | 0.0 |
| SUPERFAMILY | SSF56042 | IPR036676 | PurM-like, C-terminal domain superfamily | 177 | 347 | 1.79E-61 | |
| Pfam | PF02769 | AIR synthase related protein, C-terminal domain | IPR010918 | PurM-like, C-terminal domain | 183 | 346 | 1.2E-39 |
| Hamap | MF_00741 | Phosphoribosylformylglycinamidine cyclo-ligase [purM]. | IPR004733 | Phosphoribosylformylglycinamidine cyclo-ligase | 13 | 349 | 47.554 |
| TIGRFAM | TIGR00878 | purM: phosphoribosylformylglycinamidine cyclo-ligase | IPR004733 | Phosphoribosylformylglycinamidine cyclo-ligase | 13 | 340 | 5.2E-140 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.