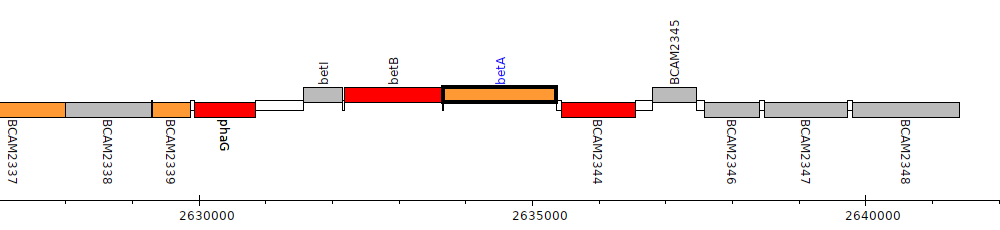
Burkholderia cenocepacia J2315, BCAM2343 (betA)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0050660 | flavin adenine dinucleotide binding |
Inferred from Sequence Model
Term mapped from: InterPro:PIRSF000137
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0016614 | oxidoreductase activity, acting on CH-OH group of donors |
Inferred from Sequence Model
Term mapped from: InterPro:PIRSF000137
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0019285 | glycine betaine biosynthetic process from choline |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR01810
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0008812 | choline dehydrogenase activity |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR01810
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0008812 | choline dehydrogenase activity |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR01810
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0050660 | flavin adenine dinucleotide binding |
Inferred from Sequence Model
Term mapped from: InterPro:PIRSF000137
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0016614 | oxidoreductase activity, acting on CH-OH group of donors |
Inferred from Sequence Model
Term mapped from: InterPro:PIRSF000137
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0055114 | oxidation-reduction process |
Inferred from Sequence Model
Term mapped from: InterPro:PIRSF000137
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0019285 | glycine betaine biosynthetic process from choline |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR01810
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0055114 | oxidation-reduction process |
Inferred from Sequence Model
Term mapped from: InterPro:PIRSF000137
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | bcj00260 | Glycine, serine and threonine metabolism | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG (InterPro) | 00260 | Glycine, serine and threonine metabolism | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
|
| MetaCyc | choline degradation IV | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
||
| KEGG | bcj01100 | Metabolic pathways | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| MetaCyc | glycine betaine biosynthesis II (Gram-positive bacteria) | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| ProSitePatterns | PS00623 | GMC oxidoreductases signature 1. | IPR000172 | Glucose-methanol-choline oxidoreductase, N-terminal | 85 | 108 | - |
| TIGRFAM | TIGR01810 | betA: choline dehydrogenase | IPR011533 | Oxygen-dependent choline dehydrogenase | 7 | 538 | 8.6E-280 |
| Hamap | MF_00750 | Oxygen-dependent choline dehydrogenase [betA]. | IPR011533 | Oxygen-dependent choline dehydrogenase | 4 | 555 | 60.4 |
| SUPERFAMILY | SSF54373 | 296 | 478 | 1.47E-61 | |||
| Gene3D | G3DSA:4.10.450.10 | IPR027424 | Glucose Oxidase, domain 2 | 39 | 202 | 4.8E-175 | |
| Gene3D | G3DSA:3.50.50.60 | IPR036188 | FAD/NAD(P)-binding domain superfamily | 5 | 532 | 4.8E-175 | |
| Gene3D | G3DSA:3.30.410.40 | 155 | 475 | 4.8E-175 | |||
| PIRSF | PIRSF000137 | IPR012132 | Glucose-methanol-choline oxidoreductase | 1 | 542 | 8.0E-142 | |
| Pfam | PF00732 | GMC oxidoreductase | IPR000172 | Glucose-methanol-choline oxidoreductase, N-terminal | 6 | 301 | 1.7E-99 |
| Pfam | PF05199 | GMC oxidoreductase | IPR007867 | Glucose-methanol-choline oxidoreductase, C-terminal | 394 | 528 | 1.6E-38 |
| Gene3D | G3DSA:1.10.1220.110 | 100 | 154 | 4.8E-175 | |||
| SUPERFAMILY | SSF51905 | IPR036188 | FAD/NAD(P)-binding domain superfamily | 1 | 536 | 1.44E-89 | |
| ProSitePatterns | PS00624 | GMC oxidoreductases signature 2. | IPR000172 | Glucose-methanol-choline oxidoreductase, N-terminal | 260 | 274 | - |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.