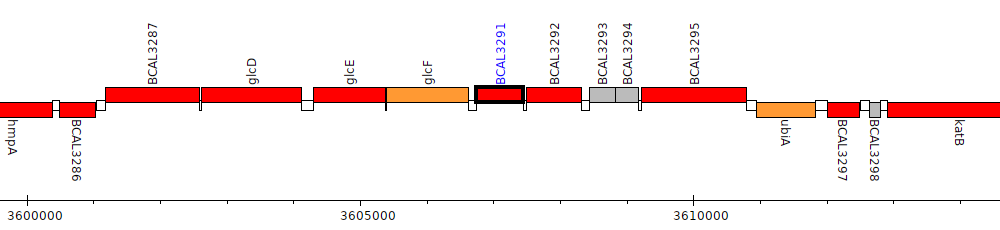
Burkholderia cenocepacia J2315, BCAL3291
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0030170 | pyridoxal phosphate binding |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR00044
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG (InterPro) | 00473 | D-Alanine metabolism | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
|
| MetaCyc | anaerobic energy metabolism (invertebrates, cytosol) | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
||
| MetaCyc | ansatrienin biosynthesis | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| SUPERFAMILY | SSF51419 | IPR029066 | PLP-binding barrel | 1 | 226 | 3.93E-82 | |
| PIRSF | PIRSF004848 | IPR011078 | Pyridoxal phosphate homeostasis protein | 1 | 232 | 3.0E-75 | |
| TIGRFAM | TIGR00044 | TIGR00044: pyridoxal phosphate enzyme, YggS family | IPR011078 | Pyridoxal phosphate homeostasis protein | 1 | 228 | 1.3E-76 |
| Hamap | MF_02087 | Pyridoxal phosphate homeostasis protein. | IPR011078 | Pyridoxal phosphate homeostasis protein | 6 | 227 | 33.109 |
| CDD | cd06824 | PLPDE_III_Yggs_like | 26 | 227 | 1.20258E-133 | ||
| Gene3D | G3DSA:3.20.20.10 | IPR029066 | PLP-binding barrel | 1 | 232 | 4.2E-89 | |
| Pfam | PF01168 | Alanine racemase, N-terminal domain | IPR001608 | Alanine racemase, N-terminal | 8 | 228 | 3.6E-34 |
| ProSitePatterns | PS01211 | Uncharacterized protein family UPF0001 signature. | IPR011078 | Pyridoxal phosphate homeostasis protein | 78 | 92 | - |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.