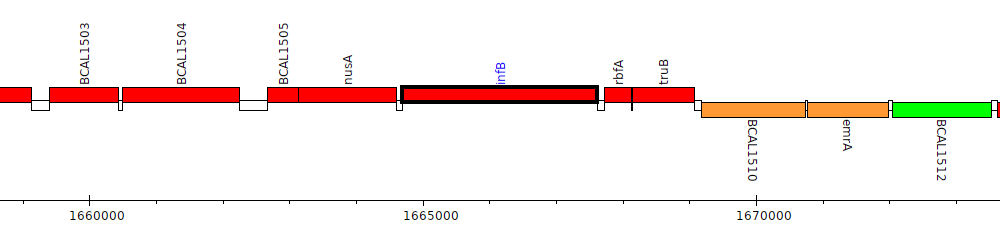
Burkholderia cenocepacia J2315, BCAL1507 (infB)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0003924 | GTPase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PS51722
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0006413 | translational initiation |
Inferred from Sequence Model
Term mapped from: InterPro:PS01176
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0003743 | translation initiation factor activity |
Inferred from Sequence Model
Term mapped from: InterPro:PS01176
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0005525 | GTP binding |
Inferred from Sequence Model
Term mapped from: InterPro:PS01176
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| TIGRFAM | TIGR00231 | small_GTP: small GTP-binding protein domain | IPR005225 | Small GTP-binding protein domain | 474 | 630 | 2.9E-31 |
| Pfam | PF11987 | Translation-initiation factor 2 | IPR023115 | Translation initiation factor IF- 2, domain 3 | 757 | 861 | 5.6E-38 |
| CDD | cd03692 | mtIF2_IVc | 878 | 961 | 6.87713E-43 | ||
| Gene3D | G3DSA:3.40.50.10050 | IPR036925 | Translation initiation factor IF-2, domain 3 superfamily | 765 | 872 | 1.2E-38 | |
| Pfam | PF04760 | Translation initiation factor IF-2, N-terminal region | IPR006847 | Translation initiation factor IF-2, N-terminal | 4 | 51 | 1.1E-5 |
| CDD | cd03702 | IF2_mtIF2_II | 646 | 740 | 9.12008E-48 | ||
| MobiDBLite | mobidb-lite | consensus disorder prediction | 340 | 354 | - | ||
| CDD | cd01887 | IF2_eIF5B | 474 | 637 | 1.31672E-100 | ||
| Gene3D | G3DSA:2.40.30.10 | 643 | 761 | 3.4E-39 | |||
| MobiDBLite | mobidb-lite | consensus disorder prediction | 49 | 85 | - | ||
| Coils | Coil | 217 | 257 | - | |||
| Gene3D | G3DSA:3.40.50.300 | 469 | 642 | 4.2E-71 | |||
| SUPERFAMILY | SSF50447 | IPR009000 | Translation protein, beta-barrel domain superfamily | 873 | 969 | 6.87E-33 | |
| SUPERFAMILY | SSF50447 | IPR009000 | Translation protein, beta-barrel domain superfamily | 644 | 726 | 1.72E-25 | |
| SUPERFAMILY | SSF52156 | IPR036925 | Translation initiation factor IF-2, domain 3 superfamily | 749 | 878 | 2.75E-38 | |
| SUPERFAMILY | SSF46955 | IPR009061 | Putative DNA-binding domain superfamily | 4 | 51 | 5.86E-10 | |
| MobiDBLite | mobidb-lite | consensus disorder prediction | 214 | 262 | - | ||
| MobiDBLite | mobidb-lite | consensus disorder prediction | 192 | 208 | - | ||
| SUPERFAMILY | SSF52540 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase | 468 | 640 | 1.7E-52 | |
| ProSiteProfiles | PS51722 | Translational (tr)-type guanine nucleotide-binding (G) domain profile. | IPR000795 | Transcription factor, GTP-binding domain | 471 | 640 | 53.761 |
| TIGRFAM | TIGR00487 | IF-2: translation initiation factor IF-2 | IPR000178 | Translation initiation factor aIF-2, bacterial-like | 389 | 971 | 2.5E-263 |
| MobiDBLite | mobidb-lite | consensus disorder prediction | 49 | 77 | - | ||
| MobiDBLite | mobidb-lite | consensus disorder prediction | 99 | 386 | - | ||
| Coils | Coil | 119 | 190 | - | |||
| Gene3D | G3DSA:3.30.56.50 | 3 | 51 | 1.8E-5 | |||
| Hamap | MF_00100_B | Translation initiation factor IF-2 [infB]. | IPR000178 | Translation initiation factor aIF-2, bacterial-like | 3 | 969 | 17.597 |
| ProSitePatterns | PS01176 | Initiation factor 2 signature. | IPR000178 | Translation initiation factor aIF-2, bacterial-like | 922 | 944 | - |
| MobiDBLite | mobidb-lite | consensus disorder prediction | 116 | 179 | - | ||
| Pfam | PF04760 | Translation initiation factor IF-2, N-terminal region | IPR006847 | Translation initiation factor IF-2, N-terminal | 394 | 444 | 1.6E-13 |
| Pfam | PF00009 | Elongation factor Tu GTP binding domain | IPR000795 | Transcription factor, GTP-binding domain | 476 | 631 | 5.6E-35 |
| Gene3D | G3DSA:2.40.30.10 | 873 | 970 | 6.9E-33 | |||
| Pfam | PF08364 | Bacterial translation initiation factor IF-2 associated region | IPR013575 | Initiation factor 2 associated domain, bacterial | 61 | 99 | 1.3E-15 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.