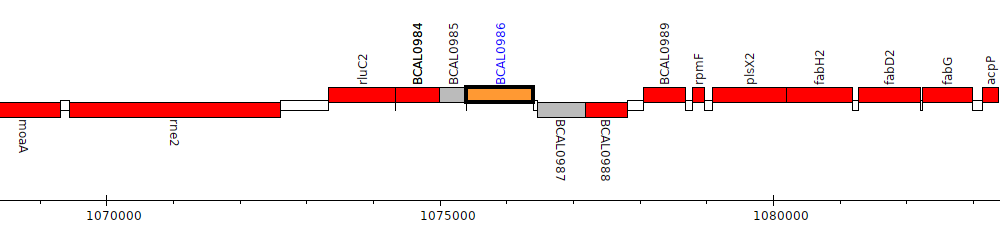
Burkholderia cenocepacia J2315, BCAL0986
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0006508 | proteolysis |
Inferred from Sequence Model
Term mapped from: InterPro:PR00127
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0008233 | peptidase activity |
Inferred from Sequence Model
Term mapped from: InterPro:cd07023
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0004252 | serine-type endopeptidase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PR00127
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| SUPERFAMILY | SSF52096 | IPR029045 | ClpP/crotonase-like domain superfamily | 70 | 277 | 2.08E-42 | |
| Pfam | PF01343 | Peptidase family S49 | IPR002142 | Peptidase S49 | 144 | 284 | 1.2E-33 |
| CDD | cd07023 | S49_Sppa_N_C | IPR002142 | Peptidase S49 | 77 | 280 | 9.84841E-84 |
| PRINTS | PR00127 | Clp protease catalytic subunit P signature | IPR001907 | ATP-dependent Clp protease proteolytic subunit | 113 | 133 | 9.1E-5 |
| Gene3D | G3DSA:3.40.1750.20 | 181 | 247 | 6.0E-62 | |||
| PRINTS | PR00127 | Clp protease catalytic subunit P signature | IPR001907 | ATP-dependent Clp protease proteolytic subunit | 254 | 273 | 9.1E-5 |
| Gene3D | G3DSA:3.90.226.10 | 81 | 281 | 6.0E-62 | |||
| PRINTS | PR00127 | Clp protease catalytic subunit P signature | IPR001907 | ATP-dependent Clp protease proteolytic subunit | 149 | 166 | 9.1E-5 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.