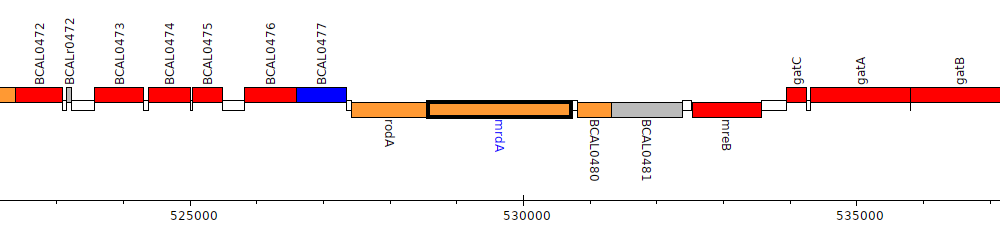
Burkholderia cenocepacia J2315, BCAL0479 (mrdA)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0009252 | peptidoglycan biosynthetic process |
Inferred from Sequence Model
Term mapped from: InterPro:MF_02081
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0009002 | serine-type D-Ala-D-Ala carboxypeptidase activity |
Inferred from Sequence Model
Term mapped from: InterPro:MF_02081
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Cellular Component | GO:0016020 | membrane |
Inferred from Sequence Model
Term mapped from: InterPro:MF_02081
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0008658 | penicillin binding |
Inferred from Sequence Model
Term mapped from: InterPro:MF_02081
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | bcj01501 | beta-Lactam resistance | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG (InterPro) | 00550 | Peptidoglycan biosynthesis | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
|
| KEGG | bcj00550 | Peptidoglycan biosynthesis | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| MetaCyc | peptidoglycan biosynthesis II (staphylococci) | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
||
| MetaCyc | peptidoglycan biosynthesis IV (Enterococcus faecium) | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Gene3D | G3DSA:3.30.1390.30 | 82 | 158 | 8.4E-41 | |||
| TIGRFAM | TIGR03423 | pbp2_mrdA: penicillin-binding protein 2 | IPR017790 | Penicillin-binding protein 2 | 17 | 627 | 1.3E-231 |
| SUPERFAMILY | SSF56601 | IPR012338 | Beta-lactamase/transpeptidase-like | 266 | 627 | 5.05E-102 | |
| Hamap | MF_02081 | Peptidoglycan D,D-transpeptidase MrdA [mrdA]. | IPR017790 | Penicillin-binding protein 2 | 3 | 631 | 49.958 |
| SUPERFAMILY | SSF56519 | IPR036138 | Penicillin-binding protein, dimerisation domain superfamily | 53 | 265 | 4.05E-46 | |
| Gene3D | G3DSA:3.90.1310.10 | 61 | 260 | 8.4E-41 | |||
| Pfam | PF00905 | Penicillin binding protein transpeptidase domain | IPR001460 | Penicillin-binding protein, transpeptidase | 283 | 624 | 2.1E-69 |
| MobiDBLite | mobidb-lite | consensus disorder prediction | 676 | 710 | - | ||
| Pfam | PF03717 | Penicillin-binding Protein dimerisation domain | IPR005311 | Penicillin-binding protein, dimerisation domain | 61 | 251 | 3.0E-40 |
| Gene3D | G3DSA:3.40.710.10 | 261 | 638 | 1.5E-101 | |||
| MobiDBLite | mobidb-lite | consensus disorder prediction | 190 | 210 | - |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.