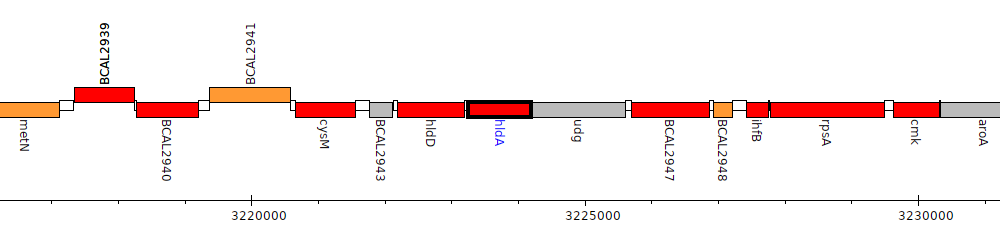
Burkholderia cenocepacia J2315, BCAL2945 (hldA)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Feature Overview
| Strain |
Burkholderia cenocepacia J2315
GCF_000009485.1|latest |
| Locus Tag |
BCAL2945
|
| NCBI Locus Tag | QU43_RS51370 |
| Name |
hldA
|
| Replicon | chromosome 1 |
| Genomic location | 3223247 - 3224197 (- strand) |
Cross-References
| RefSeq | YP_002232045.1 |
| GI | 206561280 |
| Entrez | 6933726 |
| GI | 206561280 |
| INSDC | CAR53246.1 |
| NCBI Locus Tag | QU43_RS51370 |
| NCBI Old Locus Tag | BCAL2945 |
| RefSeq | WP_006486931.1,GeneID |
| UniParc | UPI00017B9732 |
| UniProtKB Acc | B4EB35 |
| UniProtKB ID | B4EB35_BURCJ |
| UniRef100 | UniRef100_B4EB35 |
| UniRef50 | UniRef50_Q7WGU8 |
| UniRef90 | UniRef90_B4EB35 |
Product
| Feature Type | CDS |
| Coding Frame | 1 |
| Product Name |
D-beta-D-heptose 7-phosphate kinase
|
| Synonyms | |
| Evidence for Translation | |
| Charge (pH 7) | -7.36 |
| Kyte-Doolittle Hydrophobicity Value | 0.092 |
| Molecular Weight (kDa) | 34004.8 |
| Isoelectric Point (pI) | 5.24 |
Subcellular localization
| Individual Mappings | |
| Additional evidence for subcellular localization |
PDB 3D Structures
| Accession | Header | Accession Date | Compound | Source | Resolution | Method | Percent Identity |
| 4E84 | TRANSFERASE | 03/19/12 | Crystal Structure of Burkholderia cenocepacia HldA | BURKHOLDERIA CENOCEPACIA | 2.6 | X-RAY DIFFRACTION | 100.0 |
| 4E8Y | Transferase/Transferase Inhibitor | 03/20/12 | Crystal Structure of Burkholderia cenocepacia HldA in Complex with an ATP-competitive Inhibitor | BURKHOLDERIA CENOCEPACIA | 2.6 | X-RAY DIFFRACTION | 100.0 |
| 4E8W | Transferase/Transferase Inhibitor | 03/20/12 | Crystal Structure of Burkholderia cenocepacia HldA in Complex with an ATP-competitive Inhibitor | BURKHOLDERIA CENOCEPACIA | 2.8654 | X-RAY DIFFRACTION | 100.0 |
| 4E8Z | Transferase/Transferase Inhibitor | 03/20/12 | Crystal Structure of Burkholderia cenocepacia HldA in Complex with an ATP-competitive Inhibitor | BURKHOLDERIA CENOCEPACIA | 3.05 | X-RAY DIFFRACTION | 100.0 |
Virulence Evidence
| Source 1 | |
| Database | BGD |
| Category | Lipopolysaccharide core region |
| Evidence |
ECO:0000316
genetic interaction evidence used in manual assertion
|
| Host Organism | Rattus norvegicus |
| Notes | hldA-/hdlD- mutant does not survive in the rat agar bead model of chronic lung infection |
| Reference | 16513737 |
Pathogen Association Analysis
| Results |
Common
Found in both pathogen and nonpathogenic strains
Hits to this gene were found in 341 genera
|
Orthologs/Comparative Genomics
| Burkholderia Ortholog Group |
BG000894 (653 members) |
| Putative Inparalogs | None Found |
Interactions
| STRING database | Search for predicted protein-protein interactions using:
Search term: BCAL2945
Search term: hldA
Search term: D-beta-D-heptose 7-phosphate kinase
|
Human Homologs
References
|
A complete lipopolysaccharide inner core oligosaccharide is required for resistance of Burkholderia cenocepacia to antimicrobial peptides and bacterial survival in vivo.
Loutet SA, Flannagan RS, Kooi C, Sokol PA, Valvano MA
J Bacteriol 2006 Mar;188(6):2073-80
PubMed ID: 16513737
|
|
Structural-Functional Studies of Burkholderia cenocepaciad-Glycero-β-d-manno-heptose 7-Phosphate Kinase (HldA) and Characterization of Inhibitors with Antibiotic Adjuvant and Antivirulence Properties.
Lee TW, Verhey TB, Antiperovitch PA, Atamanyuk D, Desroy N, Oliveira C, Denis A, Gerusz V, Drocourt E, Loutet SA, Hamad MA, Stanetty C, Andres SN, Sugiman-Marangos S, Kosma P, Valvano MA, Moreau F, Junop MS.
J Med Chem. 2013 Jan 22. [Epub ahead of print]
PubMed ID: 23256532
|