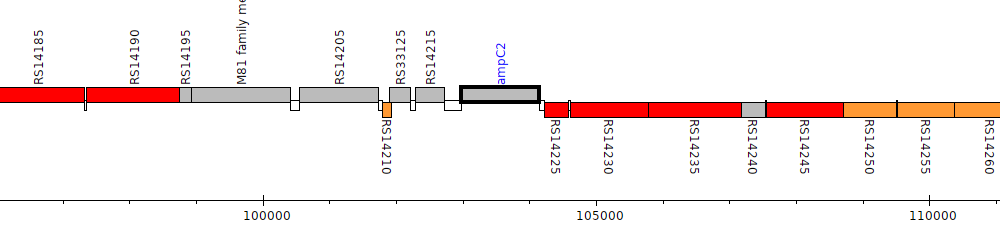
Burkholderia multivorans AU19564, C6Q10_RS14220 (ampC2)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Cellular Component | GO:0030288 | outer membrane-bounded periplasmic space |
Inferred from Sequence Model
Term mapped from: InterPro:PS00336
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0017001 | antibiotic catabolic process |
Inferred from Sequence Model
Term mapped from: InterPro:PS00336
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0008800 | beta-lactamase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PS00336
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| SUPERFAMILY | SSF56601 | IPR012338 | Beta-lactamase/transpeptidase-like | 30 | 385 | 6.6E-96 | |
| ProSitePatterns | PS00336 | Beta-lactamase class-C active site. | IPR001586 | Beta-lactamase, class-C active site | 85 | 92 | - |
| Pfam | PF00144 | Beta-lactamase | IPR001466 | Beta-lactamase-related | 38 | 382 | 7.8E-77 |
| Gene3D | G3DSA:3.40.710.10 | 29 | 388 | 2.9E-140 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.