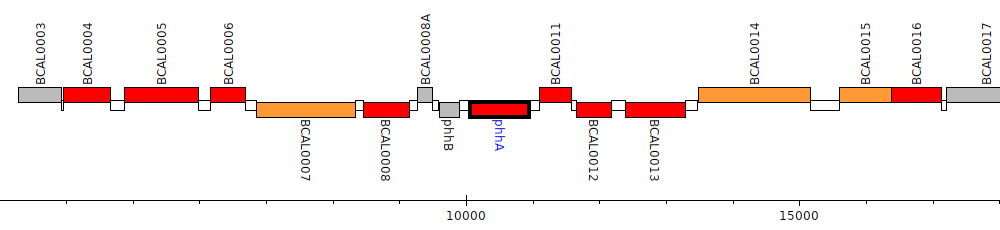
Burkholderia cenocepacia J2315, BCAL0010 (phhA)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0009072 | aromatic amino acid family metabolic process |
Inferred from Sequence Model
Term mapped from: InterPro:G3DSA:1.10.800.10
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0005506 | iron ion binding |
Inferred from Sequence Model
Term mapped from: InterPro:G3DSA:1.10.800.10
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0006559 | L-phenylalanine catabolic process |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR01267
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0004497 | monooxygenase activity |
Inferred from Sequence Model
Term mapped from: InterPro:G3DSA:1.10.800.10
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0004505 | phenylalanine 4-monooxygenase activity |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR01267
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen |
Inferred from Sequence Model
Term mapped from: InterPro:PS51410
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0055114 | oxidation-reduction process |
Inferred from Sequence Model
Term mapped from: InterPro:PS51410
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG (InterPro) | 00790 | Folate biosynthesis | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
|
| MetaCyc | L-tyrosine biosynthesis IV | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
||
| KEGG (InterPro) | 00360 | Phenylalanine metabolism | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
|
| KEGG | bcj01230 | Biosynthesis of amino acids | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | bcj01100 | Metabolic pathways | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| MetaCyc | L-phenylalanine degradation V | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
||
| KEGG (InterPro) | 00400 | Phenylalanine, tyrosine and tryptophan biosynthesis | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
|
| KEGG | bcj00400 | Phenylalanine, tyrosine and tryptophan biosynthesis | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | bcj00360 | Phenylalanine metabolism | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| ProSiteProfiles | PS51410 | Biopterin-dependent aromatic amino acid hydroxylase family profile. | IPR019774 | Aromatic amino acid hydroxylase, C-terminal | 1 | 297 | 103.106 |
| PRINTS | PR00372 | Biopterin-dependent aromatic amino acid hydroxylase signature | IPR019774 | Aromatic amino acid hydroxylase, C-terminal | 151 | 170 | 4.9E-13 |
| PRINTS | PR00372 | Biopterin-dependent aromatic amino acid hydroxylase signature | IPR019774 | Aromatic amino acid hydroxylase, C-terminal | 228 | 247 | 4.9E-13 |
| TIGRFAM | TIGR01267 | Phe4hydrox_mono: phenylalanine-4-hydroxylase | IPR005960 | Phenylalanine-4-hydroxylase, monomeric form | 22 | 268 | 7.7E-107 |
| ProSitePatterns | PS00367 | Biopterin-dependent aromatic amino acid hydroxylases signature. | IPR018301 | Aromatic amino acid hydroxylase, iron/copper binding site | 130 | 141 | - |
| SUPERFAMILY | SSF56534 | IPR036329 | Aromatic amino acid monoxygenase, C-terminal domain superfamily | 10 | 292 | 2.62E-100 | |
| CDD | cd03348 | pro_PheOH | IPR005960 | Phenylalanine-4-hydroxylase, monomeric form | 22 | 252 | 3.478E-132 |
| PRINTS | PR00372 | Biopterin-dependent aromatic amino acid hydroxylase signature | IPR019774 | Aromatic amino acid hydroxylase, C-terminal | 87 | 109 | 4.9E-13 |
| PRINTS | PR00372 | Biopterin-dependent aromatic amino acid hydroxylase signature | IPR019774 | Aromatic amino acid hydroxylase, C-terminal | 196 | 214 | 4.9E-13 |
| Pfam | PF00351 | Biopterin-dependent aromatic amino acid hydroxylase | IPR019774 | Aromatic amino acid hydroxylase, C-terminal | 32 | 249 | 1.7E-46 |
| Gene3D | G3DSA:1.10.800.10 | IPR036951 | Aromatic amino acid hydroxylase superfamily | 2 | 297 | 7.9E-106 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.