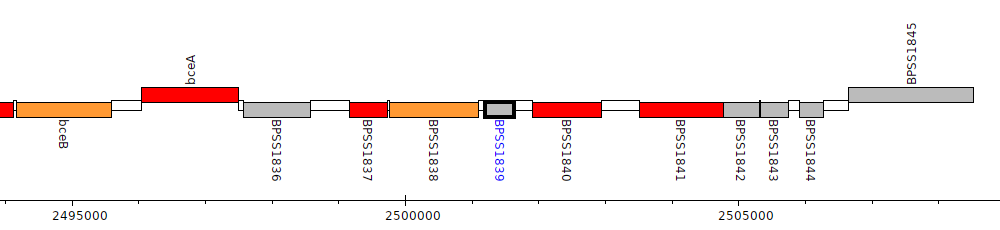
Burkholderia pseudomallei K96243, BPSS1839
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0055114 | oxidation-reduction process |
Inferred from Sequence Model
Term mapped from: InterPro:cd01041
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0016491 | oxidoreductase activity |
Inferred from Sequence Model
Term mapped from: InterPro:cd01041
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0046872 | metal ion binding |
Inferred from Sequence Model
Term mapped from: InterPro:cd01041
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Gene3D | G3DSA:1.20.1260.10 | IPR012347 | Ferritin-like | 1 | 140 | 1.0E-56 | |
| CDD | cd01041 | Rubrerythrin | IPR003251 | Rubrerythrin | 8 | 138 | 8.58469E-61 |
| SUPERFAMILY | SSF47240 | IPR009078 | Ferritin-like superfamily | 4 | 136 | 6.85E-42 | |
| ProSiteProfiles | PS50905 | Ferritin-like diiron domain profile. | IPR009040 | Ferritin-like diiron domain | 3 | 140 | 27.889 |
| Pfam | PF02915 | Rubrerythrin | IPR003251 | Rubrerythrin | 11 | 134 | 2.2E-29 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.