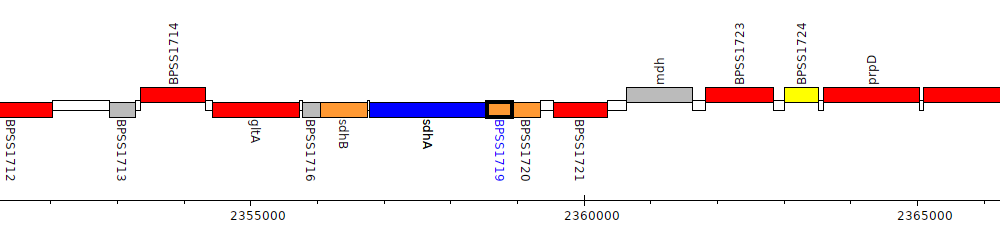
Burkholderia pseudomallei K96243, BPSS1719
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0000104 | succinate dehydrogenase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PIRSF000169
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Cellular Component | GO:0016020 | membrane |
Inferred from Sequence Model
Term mapped from: InterPro:G3DSA:1.20.1300.10
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Cellular Component | GO:0016021 | integral component of membrane |
Inferred from Sequence Model
Term mapped from: InterPro:PIRSF000169
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0020037 | heme binding |
Inferred from Sequence Model
Term mapped from: InterPro:PIRSF000169
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors |
Inferred from Sequence Model
Term mapped from: InterPro:PF01127
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0006099 | tricarboxylic acid cycle |
Inferred from Sequence Model
Term mapped from: InterPro:PIRSF000169
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | bps01200 | Carbon metabolism | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | bps01110 | Biosynthesis of secondary metabolites | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | bps00020 | Citrate cycle (TCA cycle) | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | bps00650 | Butanoate metabolism | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | bps01120 | Microbial metabolism in diverse environments | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | bps00190 | Oxidative phosphorylation | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | bps01130 | Biosynthesis of antibiotics | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | bps01100 | Metabolic pathways | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Gene3D | G3DSA:1.20.1300.10 | IPR034804 | Fumarate reductase/succinate dehydrogenase, transmembrane subunit | 10 | 122 | 2.2E-30 | |
| PIRSF | PIRSF000169 | IPR014312 | Succinate dehydrogenase, hydrophobic membrane anchor | 8 | 122 | 3.0E-40 | |
| CDD | cd03494 | SQR_TypeC_SdhD | 22 | 120 | 2.97688E-34 | ||
| SUPERFAMILY | SSF81343 | IPR034804 | Fumarate reductase/succinate dehydrogenase, transmembrane subunit | 11 | 122 | 1.57E-29 | |
| TIGRFAM | TIGR02968 | succ_dehyd_anc: succinate dehydrogenase, hydrophobic membrane anchor protein | IPR014312 | Succinate dehydrogenase, hydrophobic membrane anchor | 16 | 120 | 3.0E-37 |
| Pfam | PF01127 | Succinate dehydrogenase/Fumarate reductase transmembrane subunit | IPR000701 | Succinate dehydrogenase/fumarate reductase type B, transmembrane subunit | 2 | 106 | 5.9E-13 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.