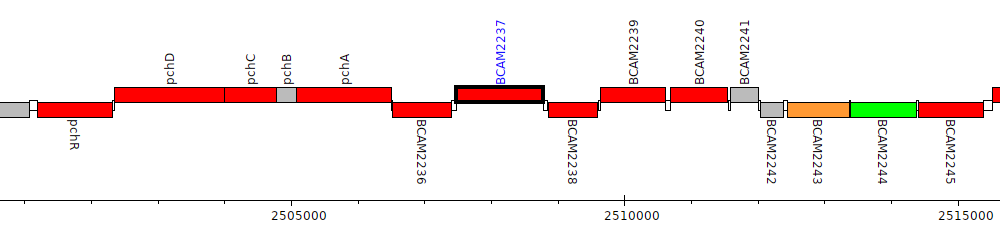
Burkholderia cenocepacia J2315, BCAM2237
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Feature Overview
| Strain |
Burkholderia cenocepacia J2315
GCF_000009485.1|latest |
| Locus Tag |
BCAM2237
|
| NCBI Locus Tag | QU43_RS65550 |
| Name |
|
| Replicon | chromosome 2 |
| Genomic location | 2507478 - 2508779 (+ strand) |
Cross-References
| RefSeq | WP_006484775.1 |
| GI | 206564072 |
| Entrez | 6929814 |
| GI | 206564072 |
| INSDC | CAR56094.1 |
| NCBI Locus Tag | QU43_RS65550 |
| NCBI Old Locus Tag | BCAM2237 |
| RefSeq | WP_006484775.1,GeneID |
| UniParc | UPI00017BA025 |
| UniProtKB Acc | B4EGG8 |
| UniProtKB ID | B4EGG8_BURCJ |
| UniRef100 | UniRef100_A0A0D1BNW0 |
| UniRef50 | UniRef50_A2QTZ7 |
| UniRef90 | UniRef90_A0A088V3D1 |
Product
| Feature Type | CDS |
| Coding Frame | 1 |
| Product Name |
putative 2,2-dialkylglycine decarboxylase
|
| Synonyms | |
| Evidence for Translation | |
| Charge (pH 7) | -4.42 |
| Kyte-Doolittle Hydrophobicity Value | 0.085 |
| Molecular Weight (kDa) | 46452.9 |
| Isoelectric Point (pI) | 5.96 |
Subcellular localization
| Individual Mappings | |
| Additional evidence for subcellular localization |
PDB 3D Structures
| Accession | Header | Accession Date | Compound | Source | Resolution | Method | Percent Identity |
| 1Z3Z | LYASE | 03/14/05 | The crystal structure of a DGD mutant: Q52A | BURKHOLDERIA CEPACIA | 2.9 | X-RAY DIFFRACTION | 94.4 |
| 1M0N | LYASE | 06/13/02 | Structure of Dialkylglycine Decarboxylase Complexed with 1-Aminocyclopentanephosphonate | BURKHOLDERIA CEPACIA | 2.2 | X-RAY DIFFRACTION | 94.7 |
| 1D7V | LYASE | 10/19/99 | CRYSTAL STRUCTURE OF THE COMPLEX OF 2,2-DIALKYLGLYCINE DECARBOXYLASE WITH NMA | BURKHOLDERIA CEPACIA | 2.8 | X-RAY DIFFRACTION | 95.2 |
| 1M0Q | LYASE | 06/13/02 | Structure of Dialkylglycine Decarboxylase Complexed with S-1-aminoethanephosphonate | BURKHOLDERIA CEPACIA | 2.0 | X-RAY DIFFRACTION | 94.7 |
| 1DKA | LYASE(DECARBOXYLASE) | 06/18/93 | DIALKYLGLYCINE DECARBOXYLASE STRUCTURE: BIFUNCTIONAL ACTIVE SITE AND ALKALI METAL BINDING SITES | BURKHOLDERIA CEPACIA | 2.6 | X-RAY DIFFRACTION | 94.7 |
| 1M0O | LYASE | 06/13/02 | Structure of Dialkylglycine Decarboxylase Complexed with 1-Amino-1-methylpropanephosphonate | BURKHOLDERIA CEPACIA | 2.4 | X-RAY DIFFRACTION | 94.7 |
| 1ZOD | LYASE | 05/12/05 | Crystal structure of dialkylglycine decarboxylase bound with cesium ion | BURKHOLDERIA CEPACIA | 1.8 | X-RAY DIFFRACTION | 94.7 |
| 1ZOB | LYASE | 05/12/05 | Crystal structure of dialkylglycine decarboxylases bound with calcium ion | BURKHOLDERIA CEPACIA | 2.75 | X-RAY DIFFRACTION | 94.7 |
| 1D7S | LYASE | 10/19/99 | CRYSTAL STRUCTURE OF THE COMPLEX OF 2,2-DIALKYLGLYCINE DECARBOXYLASE WITH DCS | BURKHOLDERIA CEPACIA | 2.05 | X-RAY DIFFRACTION | 95.2 |
| 1D7U | LYASE | 10/19/99 | Crystal structure of the complex of 2,2-dialkylglycine decarboxylase with LCS | BURKHOLDERIA CEPACIA | 1.95 | X-RAY DIFFRACTION | 95.2 |
| 2DKB | LYASE(DECARBOXYLASE) | 07/12/94 | DIALKYLGLYCINE DECARBOXYLASE STRUCTURE: BIFUNCTIONAL ACTIVE SITE AND ALKALI METAL BINDING SITES | BURKHOLDERIA CEPACIA | 2.1 | X-RAY DIFFRACTION | 94.7 |
| 1M0P | LYASE | 06/13/02 | Structure of Dialkylglycine Decarboxylase Complexed with 1-Amino-1-phenylethanephosphonate | BURKHOLDERIA CEPACIA | 2.6 | X-RAY DIFFRACTION | 94.7 |
| 1ZC9 | LYASE | 04/11/05 | The crystal structure of dialkylglycine decarboxylase complex with pyridoxamine 5-phosphate | BURKHOLDERIA CEPACIA | 2.0 | X-RAY DIFFRACTION | 94.7 |
| 1D7R | LYASE | 10/19/99 | CRYSTAL STRUCTURE OF THE COMPLEX OF 2,2-DIALKYLGLYCINE DECARBOXYLASE WITH 5PA | BURKHOLDERIA CEPACIA | 2.0 | X-RAY DIFFRACTION | 95.2 |
| 1DGE | LYASE | 06/29/94 | AN ALKALI METAL ION SIZE-DEPENDENT SWITCH IN THE ACTIVE SITE STRUCTURE OF DIALKYLGLYCINE DECARBOXYLASE | BURKHOLDERIA CEPACIA | 2.8 | X-RAY DIFFRACTION | 94.9 |
| 1DGD | LYASE | 06/29/94 | AN ALKALI METAL ION SIZE-DEPENDENT SWITCH IN THE ACTIVE SITE STRUCTURE OF DIALKYLGLYCINE DECARBOXYLASE | BURKHOLDERIA CEPACIA | 2.8 | X-RAY DIFFRACTION | 94.9 |
Pathogen Association Analysis
| Results |
Common
Found in both pathogen and nonpathogenic strains
Hits to this gene were found in 638 genera
|
Orthologs/Comparative Genomics
| Burkholderia Ortholog Group |
BG003955 (205 members) |
| Putative Inparalogs | None Found |
Interactions
| STRING database | Search for predicted protein-protein interactions using:
Search term: BCAM2237
Search term: putative 2,2-dialkylglycine decarboxylase
|
Human Homologs
|
Ensembl
97, assembly
GRCh38.p12
|
alanine--glyoxylate aminotransferase 2 [Source:HGNC Symbol;Acc:HGNC:14412]
E-value:
4.7e-47
Percent Identity:
27.6
|
References
No references are associated with this feature.